0 - The DIFFRACPlus 2006 version is necessary,
or more recent versions

1- Download the eleven P2D2 subsets in binary files compressed into zip files :
The previous P2D2 version (~100.000 entries) could be compiled as a "user database" through the PDFMAINT satellite program. However, difficulties were encountered with the P2D2 2009 update to ~1.000.000 entries. It was only found possible to cut the whole database in several parts containing close to 100.000 entries each of them :
- Zeolites SLC data : part 1, part
2, part 3, part
4 (tetrahedra only);
- Zeolites BKS data : part 1, part
2, part 3, part
4, part 5, part
6 (tetrahedra only);
- GRINSP data : part 1, zeolites
- tetrahedra, N = 4 - and other N-connected frameworks with one sort of
polyhedra - triangles (N = 3), square pyramids (N = 5) and octahedra (N
= 6), ~60.000 entries ; part 2, structure
candidates combining two kinds of polyhedra connected by corners : titanosilicates,
etc, ~100.000 entries.
PCOD/P2D2 filenumbers :
- SLC part 1 : PCOD8000001 to PCOD8080000 ; part2 : 8080001 to 8160000
; part 3 : 8160001 to 824000 ; part 4 : 8240001 to 8313568.
- BKS part 1 : PCOD9000001 to PCOD9100000 ; part 2 : 9100001 to 9200000
; part 3 : 9200001 to 9300000 ; part 4 : 93000001 to 9400000 ; part 5 :
9400001 to 9500000 ; part 6 : 9500001 to 9585139.
- GRINSP data : miscellaneous
filenumbers.
2- Unzip the 11 subfiles into the directory where is Eva.exe, generally at :
C:\DiffPlus
Example (previous name of the distribution was PCOD-3, now see SLC01 to 04, BKS01 to 06 and GRI01 to 2 for the names of the 11 subfiles) :
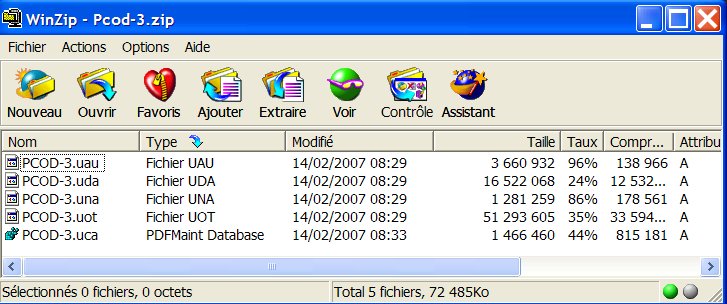
3- Start EVA, go to the settings, declare the PCOD-3.uca as being
the User Database :
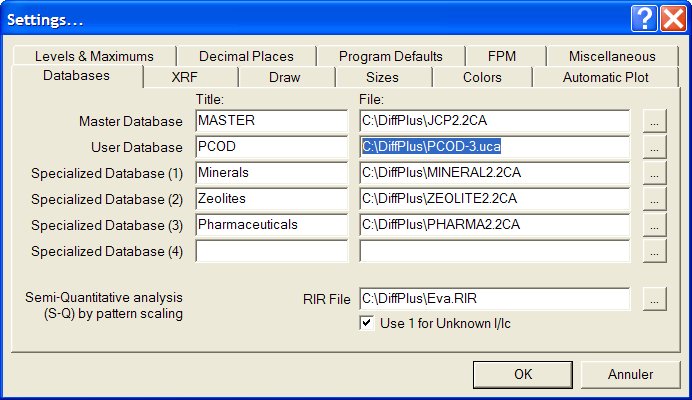
4 - When realizing identification, open the search/match box and
click to
select the option "add User Database to search" :
5 - then proceed as usual
Some (t-AlF3, K2TiSi3O9.H2O, Na4Ca4Al7F33, mordenite) were already shown in the paper Powder Diffraction 23 (2008) S5-S12, where ideal conditions (essentially less than 1% difference between the real and predicted cell parameters) for a successful search-match are described. Some of these examples are shown again here together with much more. For each example are available the raw powder pattern (Cu Kalpha) in Bruker format (.raw, which can be read by freeware like PowderX, etc) directly accessible by the EVA search-match software, screen copies of the identification (if any) by the ICDD-PDF, and by one of the P2D2 subsets (if any), with comments.
Thanks to Dr. Jens Hunger, Max Planck Institute for Chemical Physics of Solids, for sending some zeolite powder patterns.
Known zeolites Unknown zeolite (?) - Trying to solve it with the P2D2/PCOD ?
Data : t-AlF3.raw
After background and Kalpha-2 corrections, selection of a small angular
range (2-theta < 30°) which can allow for having calculated peak
positions close to experimental ones in spite of cell parameters discrpancies.
Best result from the ICDD-PDF (01-083-0717 at first place and 00-047-1659
at the fourth place, both being pertinent) :
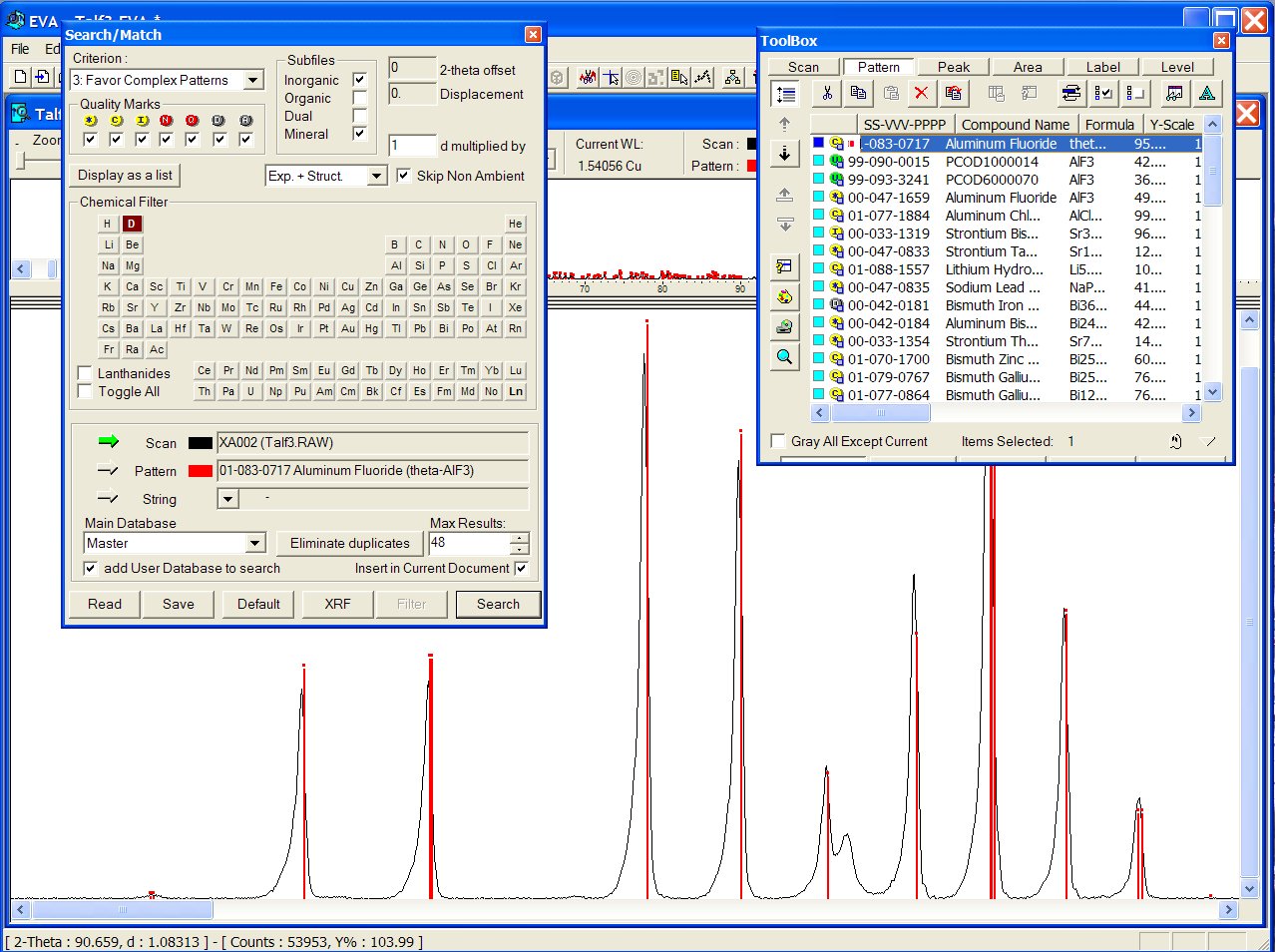
Files from the User Database are recognizable with a green U icon in the toolbox below. Correct result from the GRI01 P2D2 subset (GRINSP data), containing two times this AlF3 recent polymorph with PCOD1000014 and PCOD6000070 filenumbers, in positions 2 an 3, intermediate between the two ICDD cards :
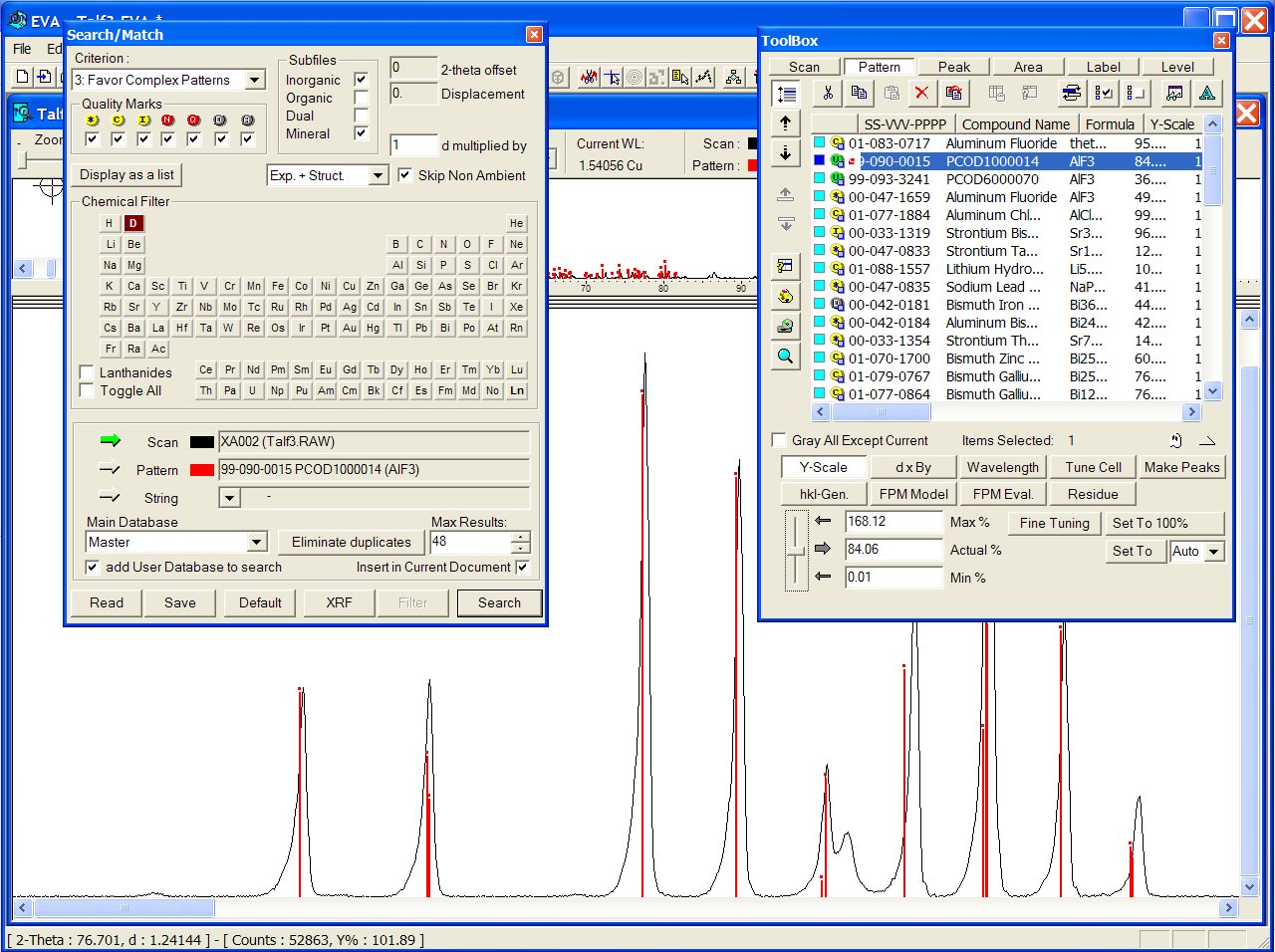
The broad unexplained peak is due to an impurity (the more usual rhombohedral
AlF3 phase).
For more about that t-AlF3 phase and the virtual MX3
compounds based on corner-sharing octahedra, see :
- Le Bail, A., Fourquet, J.L. & Bentrup, U. (1992)."t-AlF3
- crystal structure determination from X-ray powder diffraction data -
a new MX3 corner-sharing octahedra 3D network," J. Solid
State Chem. 100, 151-159.
- Le Bail, A. & Calvayrac, F. (2006)." Hypothetical AlF3
crystal structures," J. Solid State Chem.179, 3159-3166.
Data : Quartz.raw
Search-match with ICDD : Quartz-ICDD.jpg
Search-match with SLC data : Quartz-SLC04.jpg
Search-match with BKS data : Quartz-BKS02.jpg
Search-match with GRINSP data : Quartz-GRI01.jpg
If there are profusion of ICDD data matching well, one has to find manually the SLC model PCOD8299808 declared to correspond to quartz in SLC04 (cell parameters a = 4.833, c/4 = 5.346 to be compared with IZA values a = 4.916, c = 5.346, and to the real samples : a = 4.914 and c = 5.406). So, there is a discrepancy of 1.6% on a and 1.1% on c between the SLC-predicted and the real sample, then the search-match fails.
The BKS model PCOD9123753 is more successful (in spite of the fact that the BKS forcefield is not as good as the SLC) and appears in 9th position if one excludes all the deleted stuff in ICDD and PCOD3000328 in GRI01 is in 14th position.
This shows immediately the problem with a search-match approach... The powder pattern fingerprint is so accurate that predictions not much better than 1% on the (real) cell parameters will not necessarily be disclosed. In the case of real zeolites, cell variations will be associated to the atomic composition (Al substitution).
This may also suggest some improvements of the search-match software : instead of using only one processor on multi-processor machines, tests should be made on the full database but with slight cell parameter changes (+- 0.2, 0.4, 0.6 %, etc).
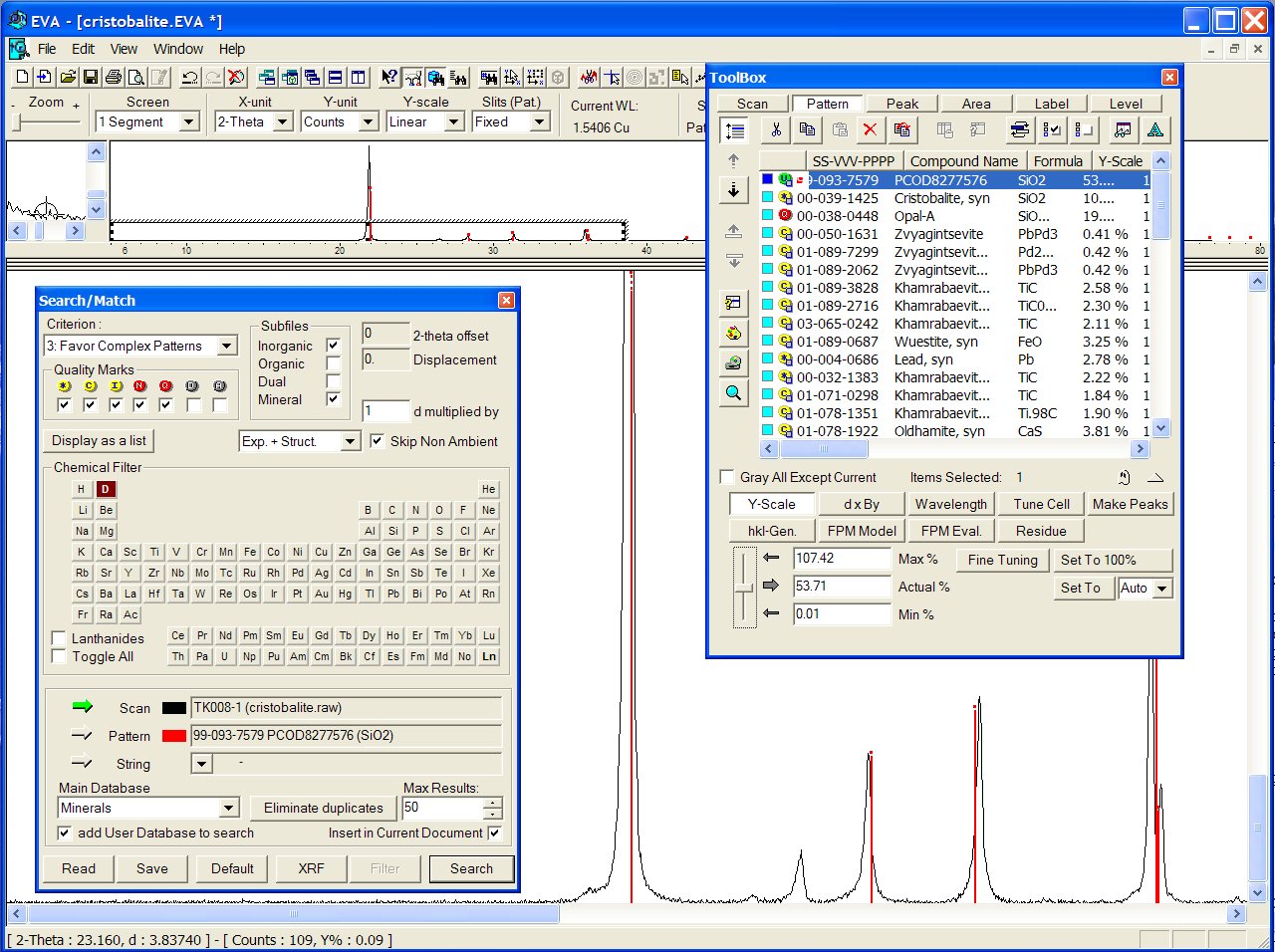
Data : cristobalite.raw
Reducing chemistry to a search on Si + O atoms, at the head of the list appears the ICDD PDF 01-076-0935 :
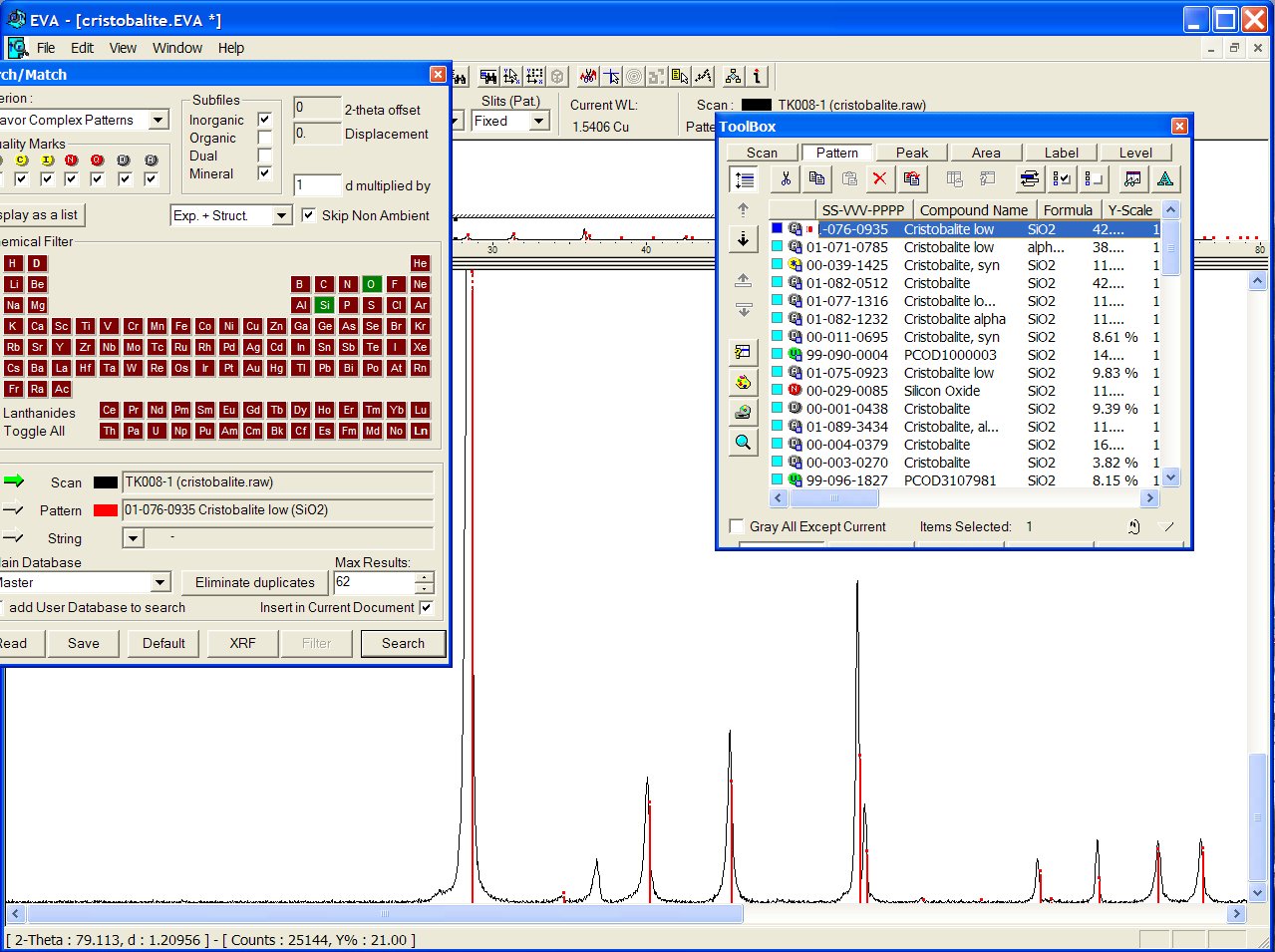
With the GRI01 P2D2 subset, the PCOD1000003 is at the 8th position among a lot of other ICDD cristobalites. It would have appeared in second position if all the "deleted" ICDD stuff had been removed.
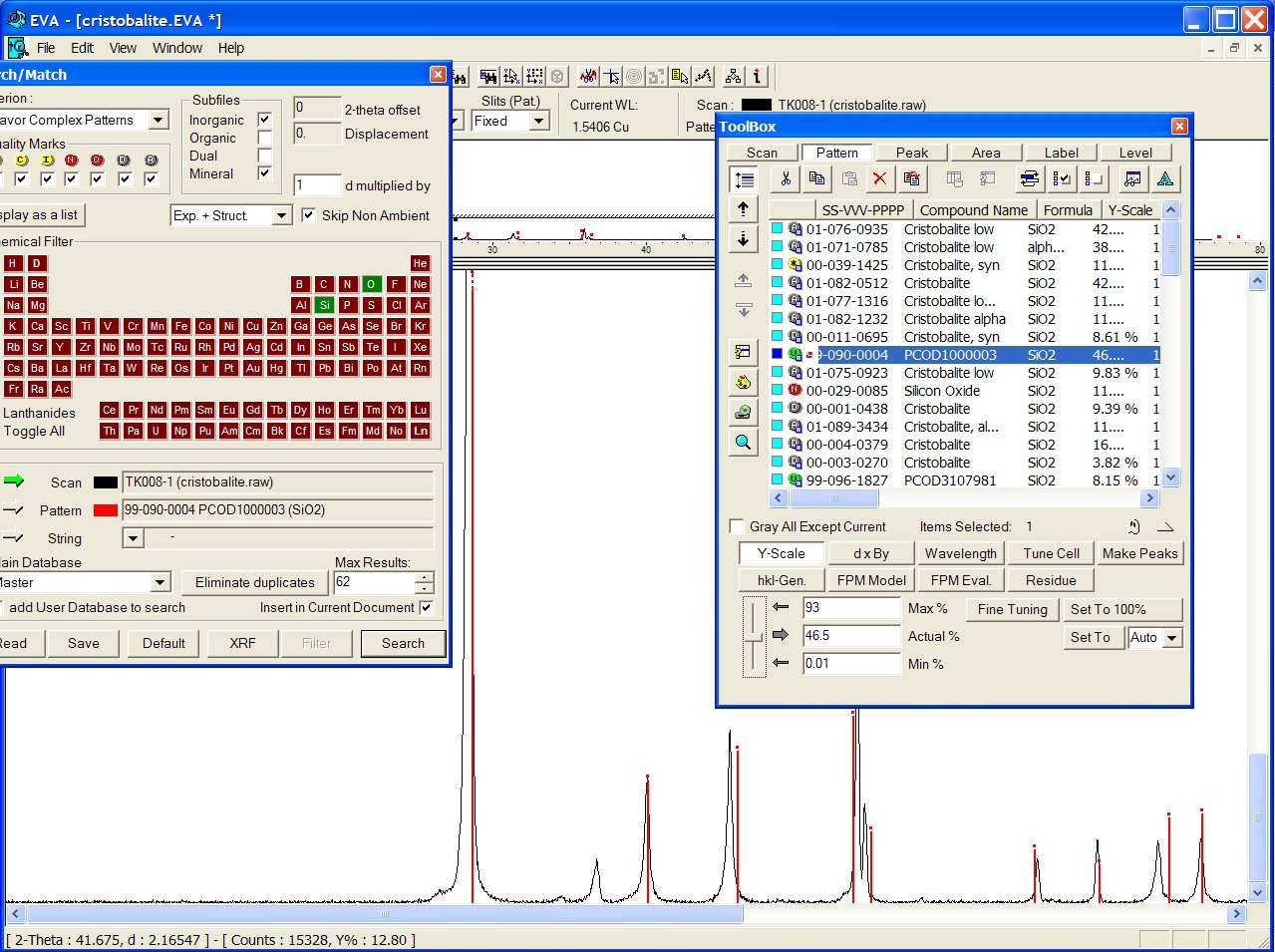
No close match was found in the BKS series, but the PCOD8277576 in SLC04 is found to match even better than ICDD, though the predicted cell is cubic instead of tetragonal :
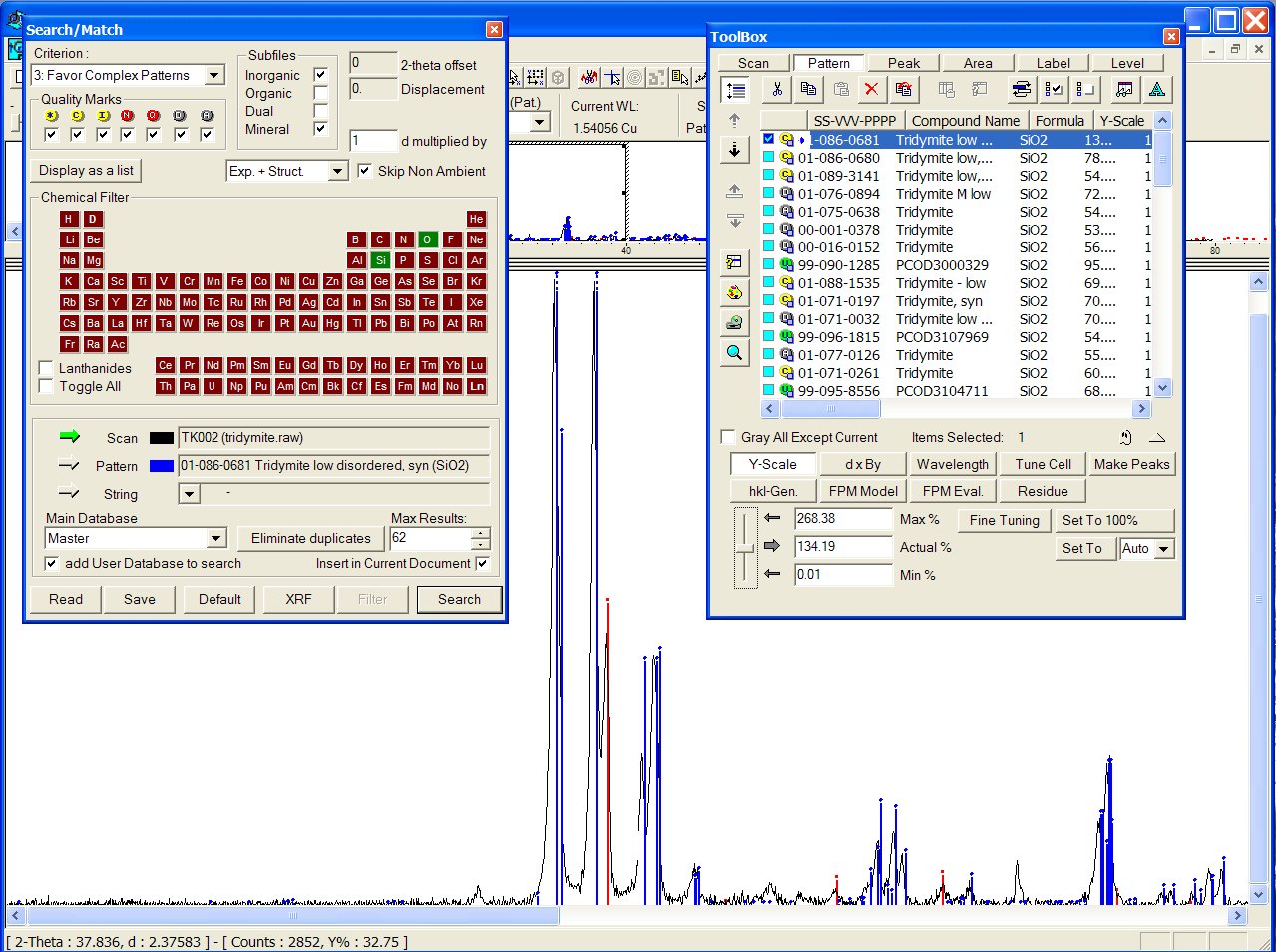
Data : tridymite.raw
Reducing chemistry to a search on Si + O atoms, at the head of the list appears the ICDD PDF 01-086-0681, and there is obviously a mixture with cristobalite (ICDD PDF 01-071-0785) :
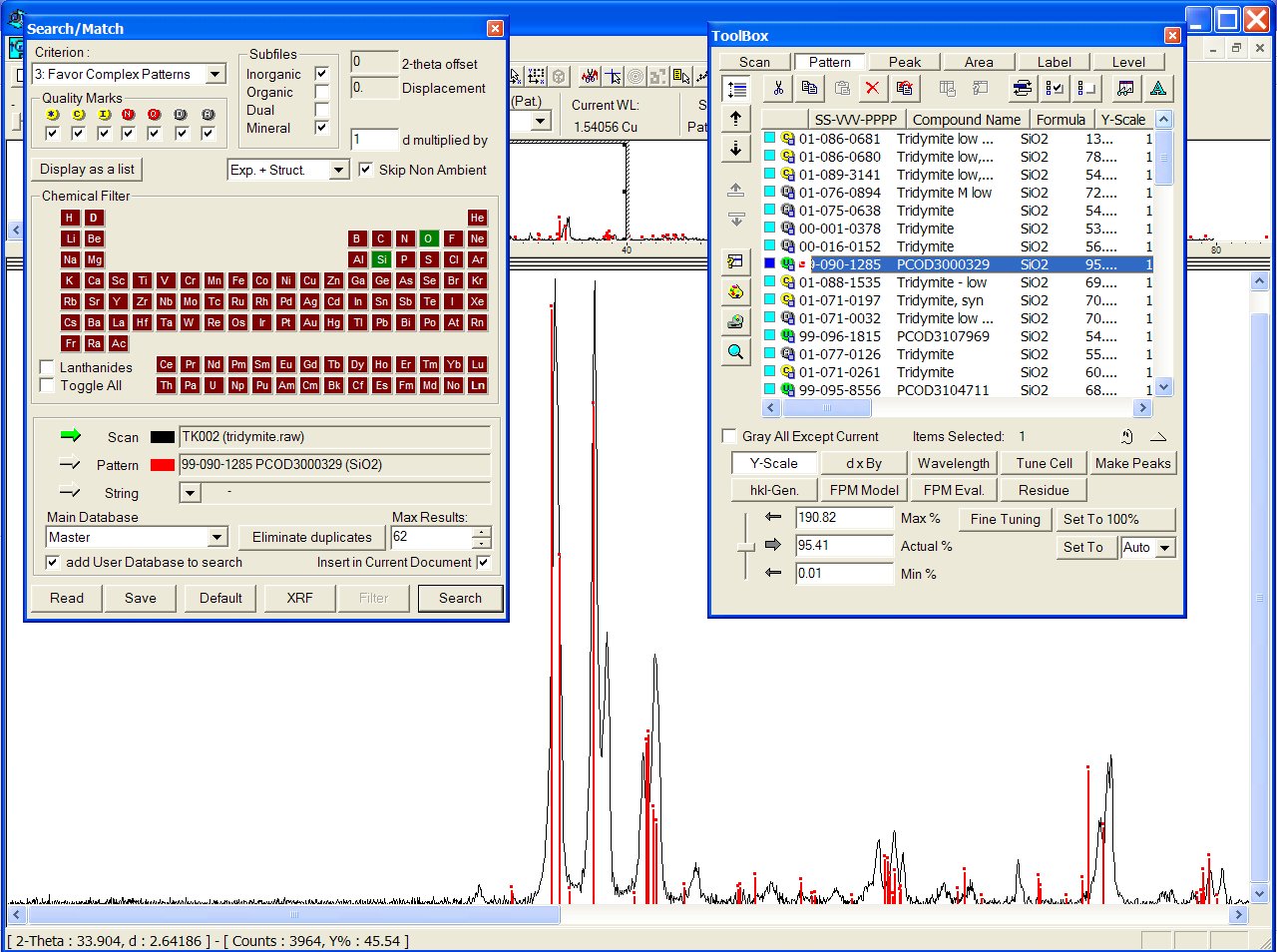
With the GRI01 P2D2 subset, the PCOD3000329 is at the 8th position among a lot of other ICDD tridymites :
Nothing is found to match very closely in the BKS series. A good match
is obtained in the SLC01 series with PCOD8057144 in third position. Verifying
the coordination sequence (CS), the tridymite framework fingerprint is
confirmed :
4 12 25 44 67 96 130 170 214 264
319 380
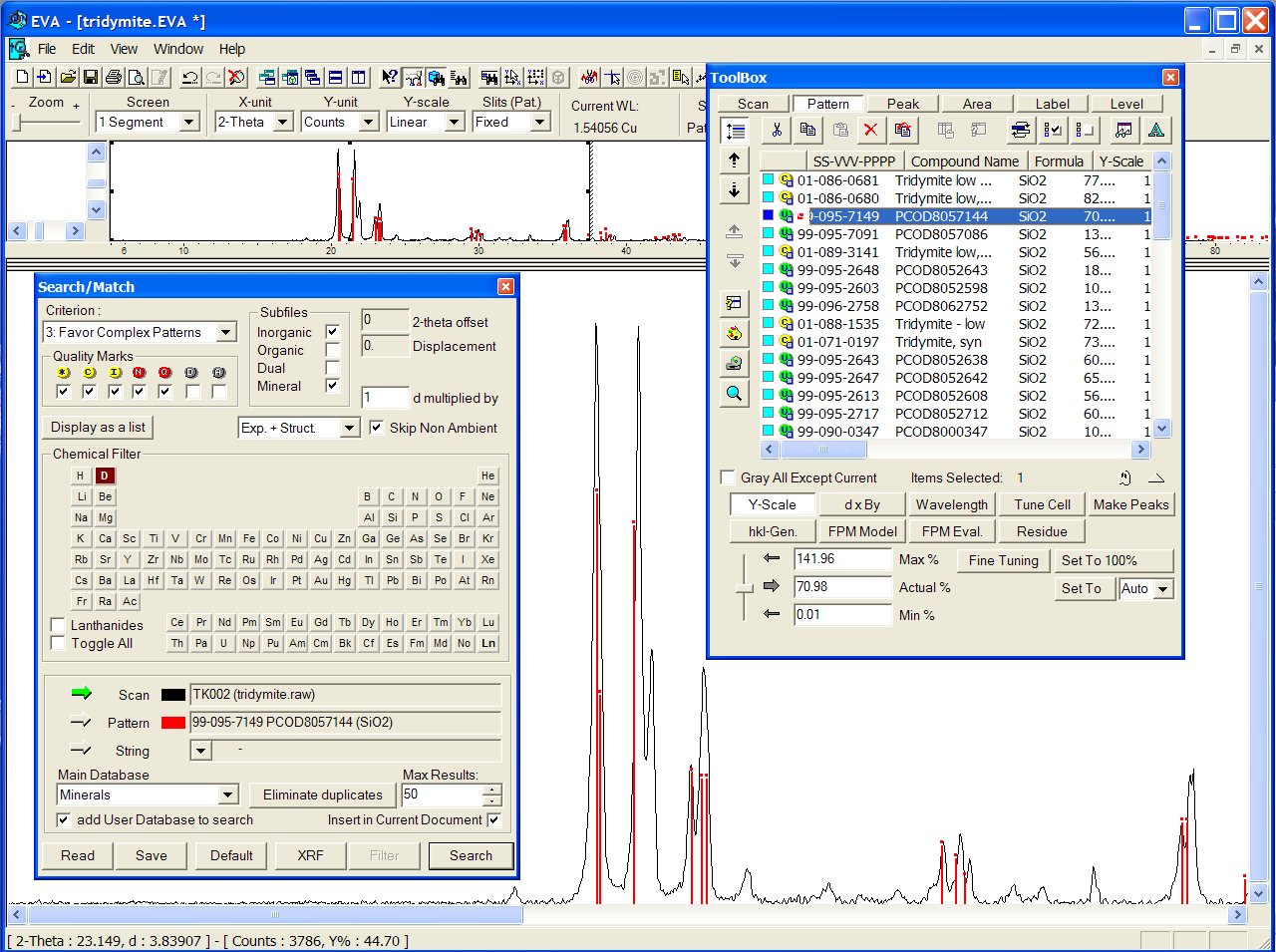
Data : Zn2P2O7.raw
ICDD identification : Zn2P2O7-ICDD.jpg
Stucture unsolved, powder pattern even not indexed.
In the P2D2 ? No match...
It could have been there from the GRINSP predictions only if the structure
was build up from ZnO5 or ZnO6 polyhedra sharing
exclusively corners with PO4 tetrahedra, but the other forms
(alpha, beta, gamma) show edge-sharing. See more on that problem in a slide
of a powerpoint file from a 2008 conference. Anyway, this is a
typical case that will have some chance to be solved today by prediction
methodologies (powder pattern with broad lines and dubious indexing).
Data : BEA.raw
ICDD identification : BEA-ICDD.jpg
Well, not a very nice sample indeed... Some match with the P2D2 : BEA-GRI01.jpg
However, the coordination sequence does not match with BEA...
Data : CHA.raw
ICDD identification : CHA-ICDD.jpg
Several data in the P2D2 were identified as being CHA by their coordination sequence. However, there is too much cell parameter discrepancies for obtaining a positive search-match. These calculated powder data are shown here by a search with their name (PCOD number) : CHA-BKS06.jpg, CHA-SLC02.jpg and CHA-GRI01.jpg
Data : FAU-NH4.raw
ICDD identification : FAU-NH4-ICDD.jpg
Same problem as for CHA above : FAU-NH4-SLC04.jpg
The idealized cubic cell parameter (Fd-3m) for FAU is a = 24.3 A (lZA), it may be 24.74 for the composition [(Ca, Mg, Na2)29(H2O)240][Al58Si134O384], for instance. The SLC prediction with a = 24.226 is close to the lZA, but the tested real sample has a = 25.128... so the search-match fails.
Data : HEU.raw
ICDD identification : HEU-ICDD.jpg
That time, the expected HEU in the P2D2 are listed in good position : HEU-BKS02.jpg and HEU-GRI01.jpg
Data : LAU.raw
ICDD identification : LAU-ICDD.jpg
Again, there is too much cell parameter discrpancies and the LAU data in the P2D2 are only obtained by a manual search : LAU-BKS02.jpg and LAU-SLC01.jpg
Data : LTA.raw
ICDD identification : LTA-ICDD.jpg
Again, a manual search is necessary : LTA-BKS06.jpg and LTA-GRI01.jpg
Data : MFI-ZSM5.raw
ICDD identification : MFI-ZSM5-ICDD.jpg
Some success is obtained with the MFI from GRINSP predictions : MFI-ZSM5-GRI01.jpg
Data : MOR.raw
ICDD identification : MOR-ICDD.jpg
Success is also abtained in the MOR case : MOR-BKS03.jpg, MOR-SLC02.jpg and MOR-GRI01.jpg
Data : NAT.raw
ICDD identification : NAT-ICDD.jpg
For NAT, the expected files are PCOD9386979 and PCOD8150320. If the
coordination sequence match, there is an enormous discrepancy between the
observed powder pattern and the model. Indeed, a framework density of 17.8
is expected, and one observes 26 for PCOD9386979. There may be some bug.
NAT-BKS04.jpg and NAT-SLC02.jpg
Data : STI.raw
ICDD identification : STI-ICDD.jpg
Matching is obtained with BKS (STI-BKS03.jpg) and GRINSP (STI-GRI01.jpg) but only manually with the SLC data (STI-SLC03.jpg).
Data : THO.raw
ICDD identification : THO-ICDD.jpg
Again, only manual fits may shown the THO recognized data (by their coordination sequence) in the P2D2 (THO-BKS04.jpg, THO-SLC02.jpg and THO-GRI01.jpg).
See the conclusions for some explanations about these deceptive results on zeolites...
This is the interesting part... Is the PCDOD/P2D2 useful for solving all those yet uncharacterized zeolites, in spite of the above deceptive results ? Maybe, if they are really zeolites with uninterrupted 3D framework with all O atoms shared by two cations in a tetrahedral coordination (not a chance for ITQ37 ansd so on because such interrupted frameworks are not predicted at all here), and if cell parameters really match, which can happen if there are not too much things into the channels... Let us see one case :
Data : LZ-200.raw
Sample from V.I. Tarasov
(1998), Institute of Silicate Chemistry, St.-Petersburg, Russia.
ICDD identification : Only one PDF card is matching (00-047-0716), unindexed, and, either the sample contains impurities or the card is uncomplete...
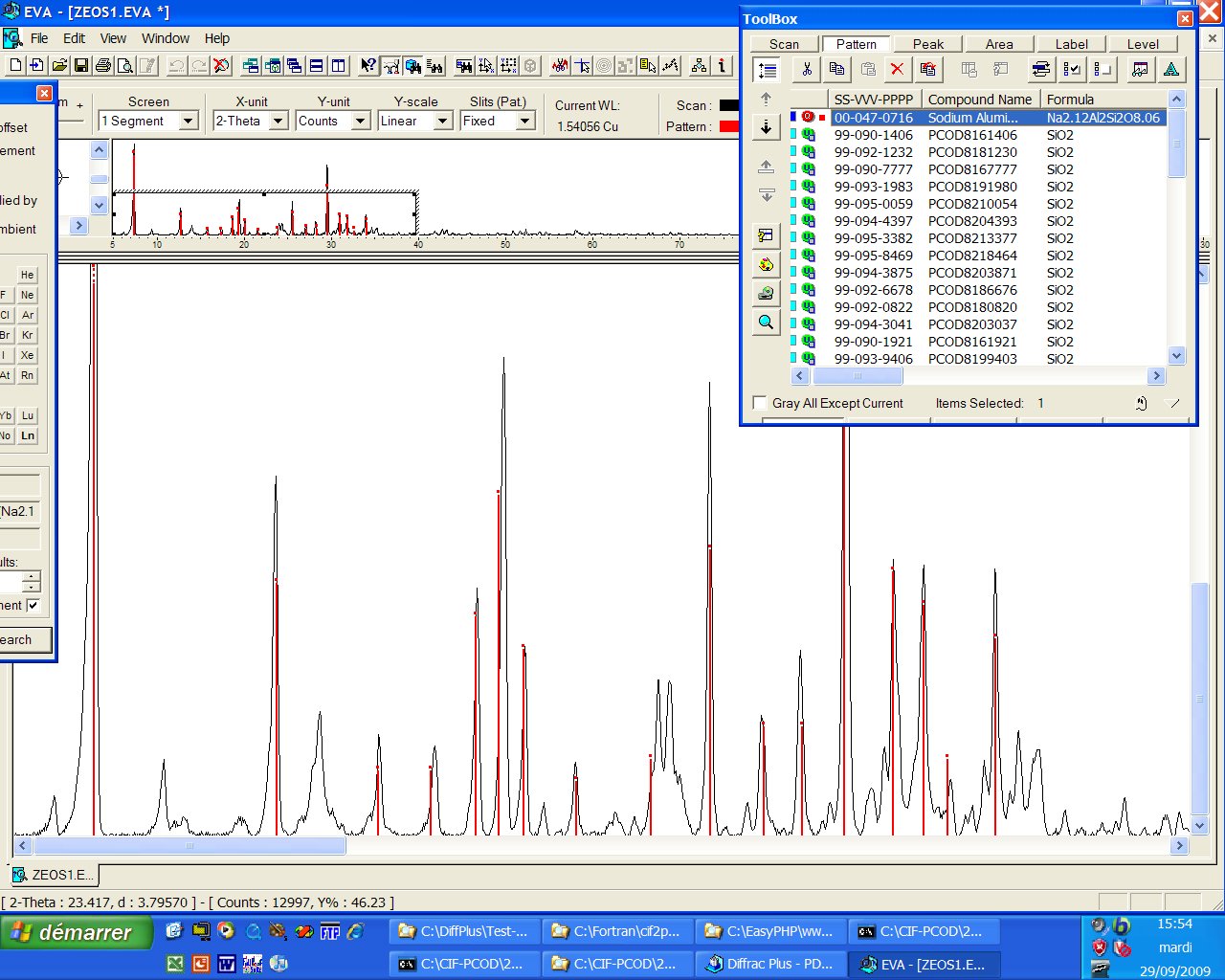
It seems that LZ-200 (also called pseudo-losod (PL), ZZ9, etc, formulation NaAlSiO4) which can be found in many patents, is possibly indexed in a hexagonal cell with a= 13.953 A, c = 9.514 A, according to the McMaille software. The impurities on the above pattern are possibly LOS, CHA, FAU (etc). Many space groups cannot be excluded, so that, after intensity extraction, no solution is coming yet (if you have a better sample, think to me...). Does prediction is suggesting something ?
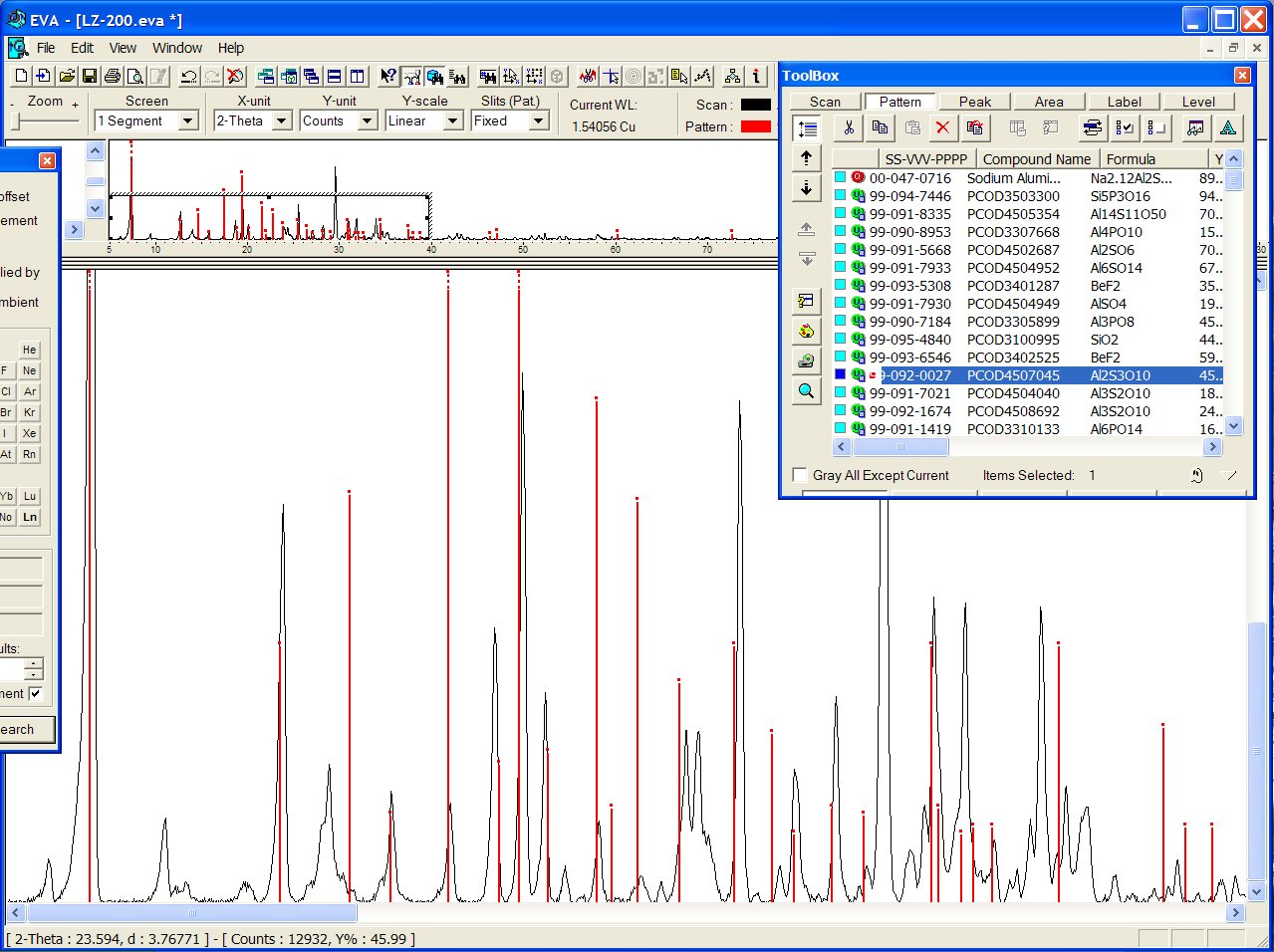
Some structure candidates have close cell parameters, for instance PCOD4507045 ("Al2S3O10"):
or PCOD3504642 ("Si2P3O10") :
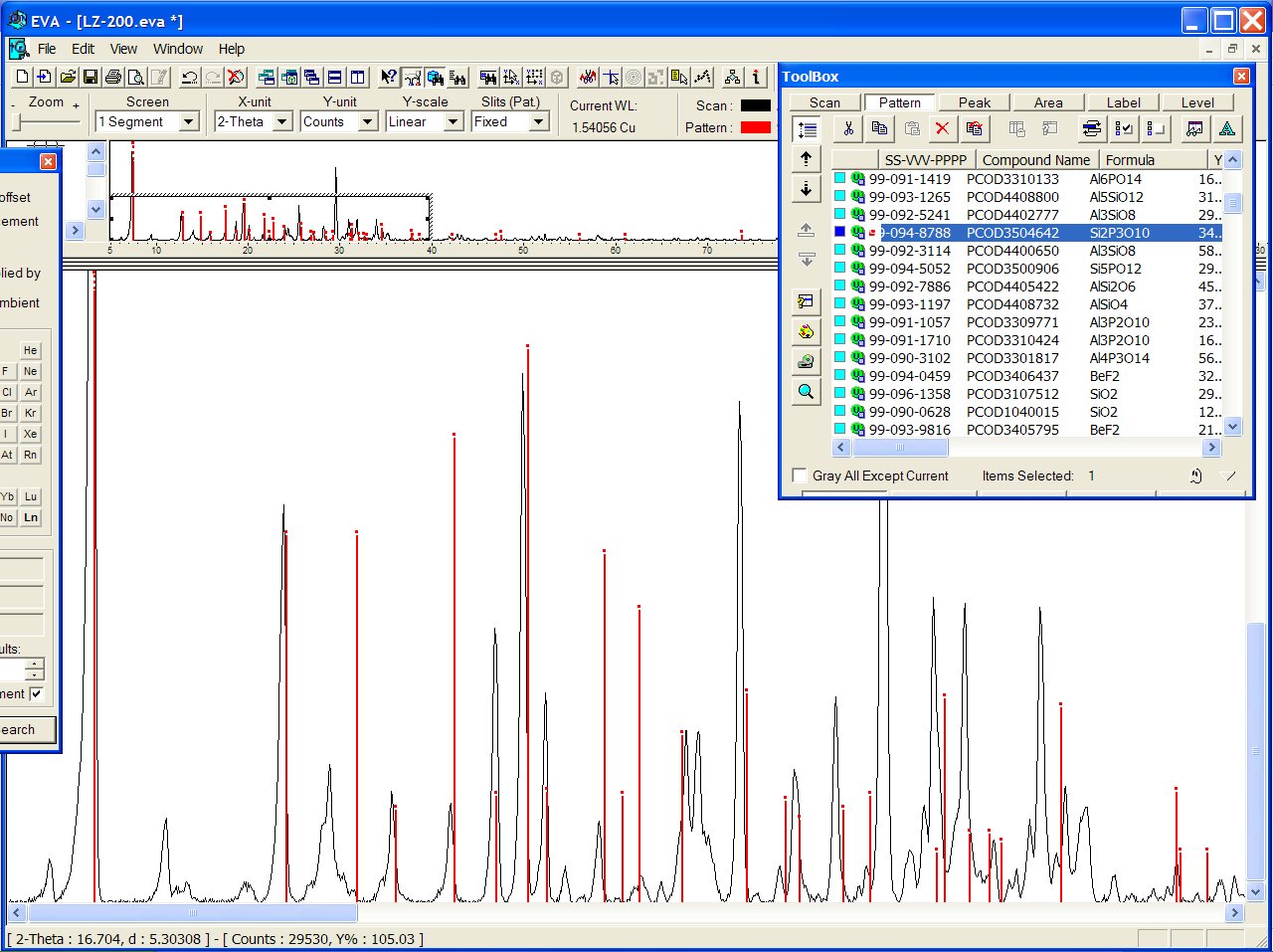
Both models have the same coordination sequence (same structure). Anyway, more work is needed before to conclude if that structure will really match :
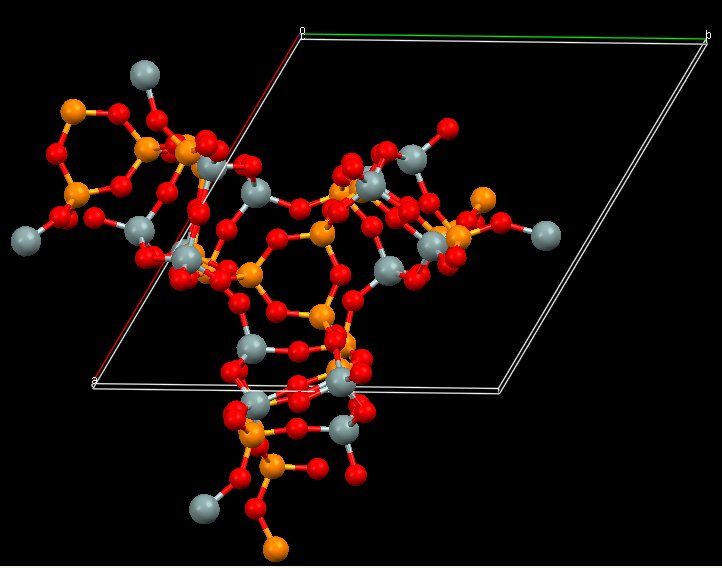
Or it may be that one, PCOD8134385, orthorhombic ?
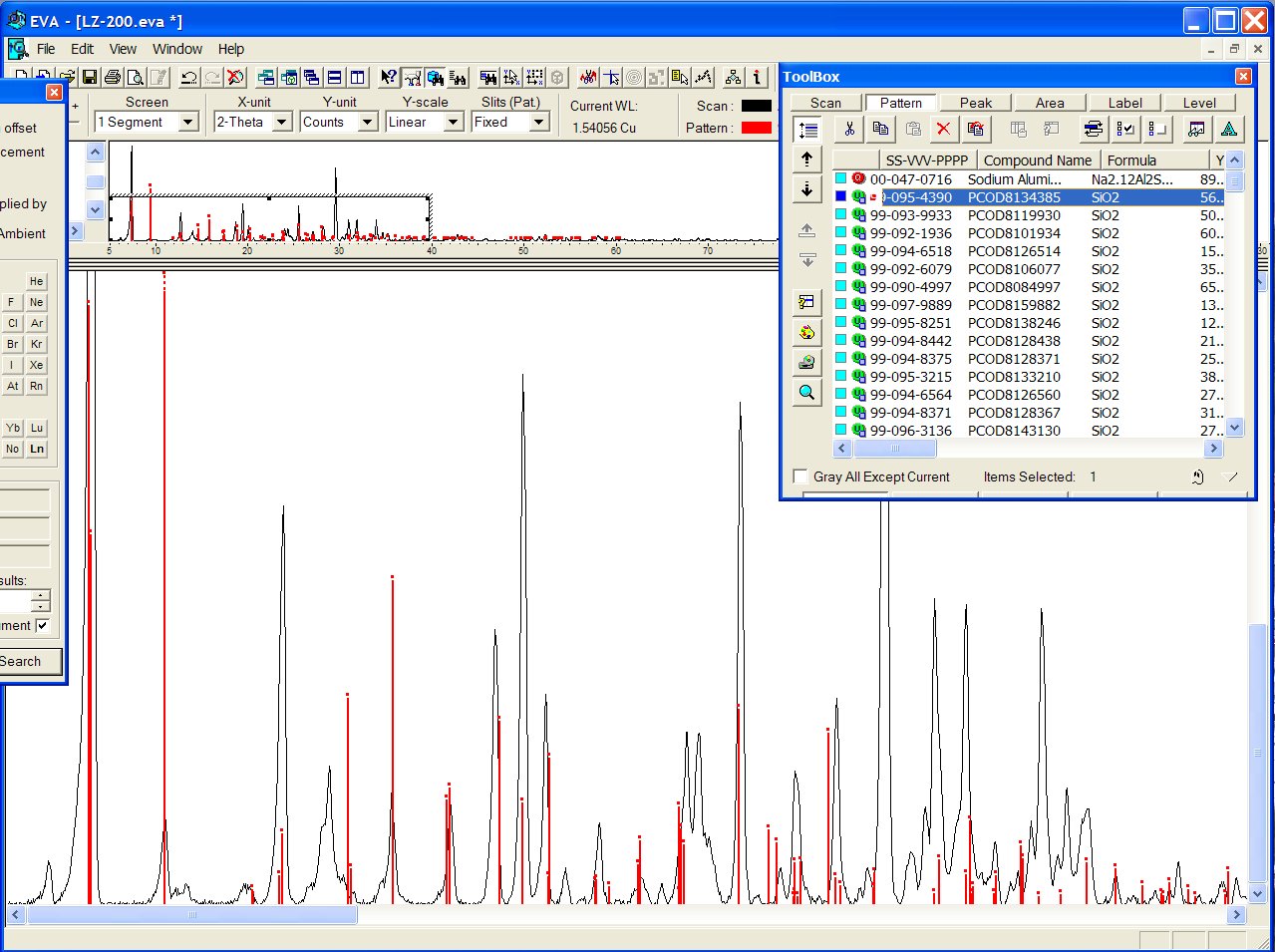
Indeed, there are 52 entries in the PCOD with cell parameters matching
at +- 0.2 A :
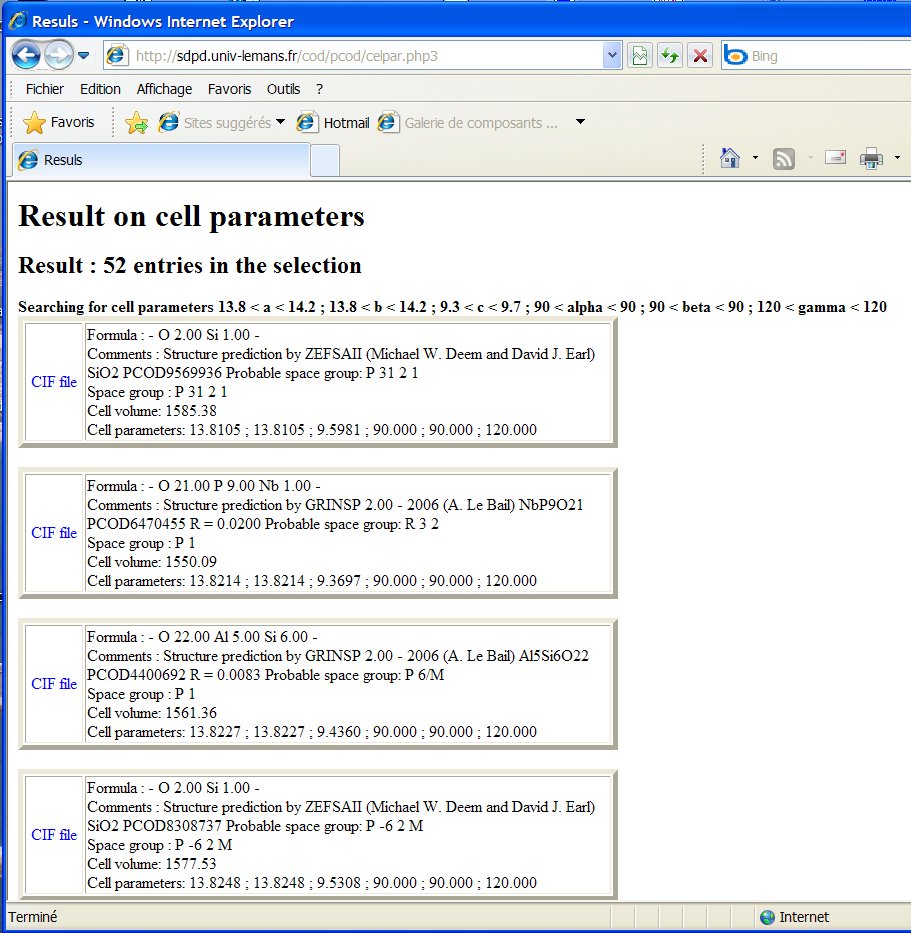
Indeed, more than these 52 should be found if the original structures predicted were checked for additional symmetries by a software like PLATON and rebuilt (for instance quartz with cx4, and many structures with ugly angles < 60° or > 130°, etc). In principle the search-match by EVA should reveal them, but the 1% tolerancy on the cell parameters is too restrictive.
Is it worth trying the PCOD/P2D2 for solving a zeolite or another problem ?
First verify if the chemical composition of your sample match with the database content or has a chance to correspond with an isostructural compound. Note that there are only framework compounds in that database (N-connected or N-N' connected 3D nets).
Even if your sample match with these requirements, it is clear from the above tests that structure prediction has progress to make... to be useful, the powder pattern fingerprint is something needing extreme accuracy (better than 1% on cell parameters)... Many known real zeolites are not correctly identified by using the P2D2 and search-match methods, even if they are declared to be well predicted... However, it must be understood that these real compounds do not have the SiO2 formula, so that the predicted cell parameters which are estimated for such a SiO2 formula cannot match well enough with a sample like [Ca9.7Na22Cl2(SO4)5.3CO3(H2O)4][Al24Si26O96] (corresponding to the zeolite AFG), for instance...
So, for unknown zeolites having resisted to crystal structure characterization (no single crystal, failure to index the powder data, etc), the interest to solve the structures is so high that the PCOD/P2D2 is really worth-trying since it remains the only chance of success. The advice is that you will increase the chances to have a correct search-match by first calcinating your sample (emptying the channels/cavities), provided the framework is not destroyed. Any new zeolite characterization will ensure you some notoriety...
Try also the P2D2 in case of titanosilicates (TiO6 octahedra and SiO4 tetrahedra connected by corners in 3D frameworks), new MX3 compounds if they are build up from corner-sharing octahedra, etc.
About other compounds (non-framework, polyhedra connections by edges, faces, etc), the improvements of the possibilities to not only predict corner-sharing polyhedra is required.
The XXI century should see CSP (Crystal Structure Prediction) becoming a more mature science, but we are now at the very beginning...