
Setting up the program files in < 10 minutes
During a recent discussion at the SDPD mailing list, the time for setting up the two files necessary for running ESPOIR was said to exceed 1 hour. It is shown below how to reduce this time to < 10 minutes in the case of a molecule location program option (this would be shorter in "scratch" mode).
Being at this stage supposes that :
- you failed at identifying your sample
as having a known structure
- you indexed the powder pattern and defined
the space group
- you extracted the structure factor amplitudes
- you failed to determine the structure
by Patterson/direct methods, etc
- you know the molecule formula and shape
1 - Retrieve the molecule atomic 3D coordinates.
Suppose that you think that your compound is a new thalidomide form. You may either try to get the molecule coordinates from the CSD database or from WebMolecules. The latter is free, so, connect to :
Click on search and the server replies very fast :
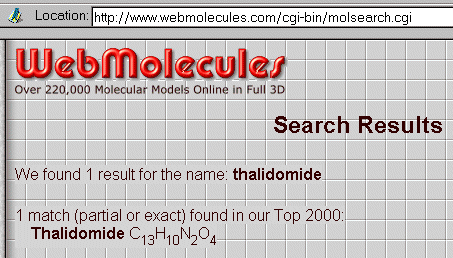
You may also use the C13H10N2O4 formula instead of the name, with a similar result :
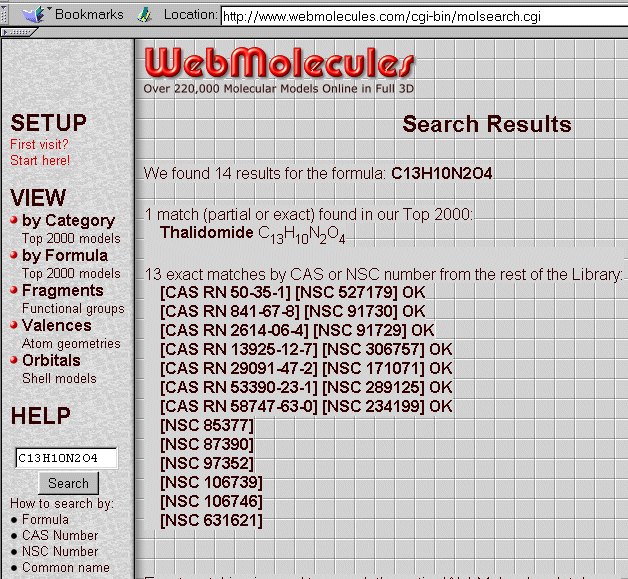
By clicking on the Webmolecules proposition, you will obtain the next screen with a drawing (your browser should be equipped with CHIME or a VRML viewer) :
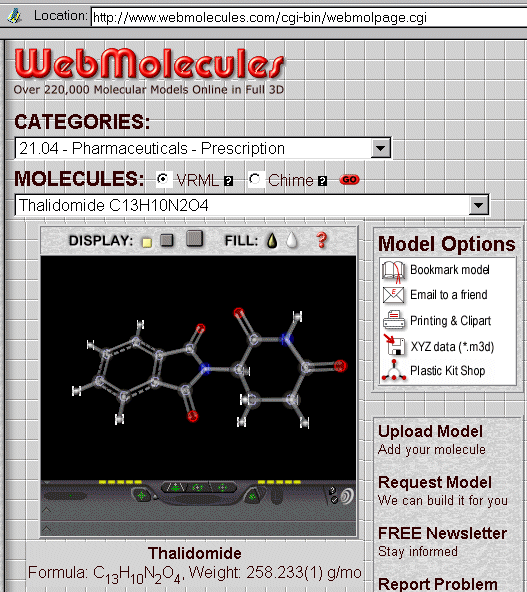
In the "Model Options" section above, click on "XYZ data (*.m3d)" and you will get the Cartesian coordinates. Save them on your computer :
STRUCTURE 1.00 1 29 31 0.000 1 Thalidomide 1 N 3 0.285 -0.041 0.232 0.000 N 2 C 3 -0.378 0.845 0.905 0.000 C 3 O 1 0.104 1.787 1.510 0.000 O 4 C 3 -0.531 -0.907 -0.290 0.000 C 5 O 1 -0.216 -1.860 -0.985 0.000 O 6 C 3 -4.058 0.603 1.110 0.000 C 7 H 1 -4.900 1.036 1.490 0.000 H 8 C 3 -4.163 -0.558 0.332 0.000 C 9 H 1 -5.081 -0.965 0.153 0.000 H 10 C 3 -3.020 -1.162 -0.200 0.000 C 11 H 1 -3.087 -2.007 -0.766 0.000 H 12 C 3 -1.795 -0.576 0.071 0.000 C 13 C 3 -1.696 0.547 0.832 0.000 C 14 C 3 -2.809 1.169 1.370 0.000 C 15 H 1 -2.720 2.011 1.939 0.000 H 16 H 1 2.119 -0.174 1.085 0.000 H 17 C 4 1.667 -0.067 0.098 0.000 C 18 H 1 1.691 -1.152 -1.752 0.000 H 19 H 1 1.736 -2.185 -0.297 0.000 H 20 C 4 2.089 -1.255 -0.743 0.000 C 21 H 1 3.970 -2.056 -1.403 0.000 H 22 H 1 4.006 -1.349 0.233 0.000 H 23 C 4 3.611 -1.239 -0.777 0.000 C 24 O 1 5.081 0.068 -1.939 0.000 O 25 C 3 4.032 0.014 -1.318 0.000 C 26 O 1 1.605 2.185 -0.462 0.000 O 27 C 3 2.204 1.120 -0.530 0.000 C 28 H 1 3.644 1.878 -1.558 0.000 H 29 N 3 3.326 1.086 -1.174 0.000 NThis has taken already 5 minutes of your precious time, if your Internet connection is slow, but these 5 minutes should not be counted in the ESPOIR files setting up which is starting now.
2- Get an appropriate previous ESPOIR .dat file and adapt it to your needs.
Your problem is a "one molecule" problem, then select a "one molecule" problem in the list of ESPOIR examples. Pyrene or RKSA1 will be fine (other examples are in the manual itself). Let us select RKSA1, copy the .dat file under another name, for instance thalid.dat, and edit it. It will look like the file below. In red is shown what you will have to change :
! title RKSA1 ! a, b, c, alpha, beta, gamma 8.7686 8.6510 10.0030 90.0 103.833 90.0 ! space group P 21 ! lambda, radiation, N of atoms, types of atoms, N of objects, ! "|Fobs|" or patterns, iprint 1.78892 4 40 4 1 1 3 ! U, V, W, step 0.10000 -0.01912 0.01836 3 ! atom names, in 8A4 C H N O Nothing to change, but verify... ! code for minimal distance contraints 0 ! maximum moves for each type of atom 10.000 10.000 10.000 10.000 ! annealing law, sigma, reject 1.0000 1.0000 0.0050 ! number of events for : print, maximum, save 10000 100000 100000 Try 250 500 for a fast test ! events for restart, rmax, ichi, number of runs 50000 0.400 2 10 ! object type and NPERM for object 1 2 4 ! number of atoms of each type in object 1 13 20 2 5 ! B overall, NOCC, NSPE for object 1 4.0 0 0 ! cell parameters, and x, y, z, occup. for object 1 !Add there the cell parameters ! and the x,y,z, occup. for your object 1 0.0 0.0 0.0 90.000 90.000 90.000 IF Cartesian 1.28649 1.51081 -1.09543 1.0 1.70731 0.39245 -0.14634 1.0 0.73658 0.09456 0.98608 1.0 -0.76365 0.23464 0.56115 1.0 -0.89683 1.61475 -0.14282 1.0 1.91698 -1.81374 0.32282 1.0 1.52353 3.87008 -1.22846 1.0 3.33699 -1.88972 0.82641 1.0 1.38502 -3.15958 -0.08199 1.0 -2.40164 -1.33363 -0.49612 1.0 -2.53843 -2.41550 -1.53132 1.0 -1.53043 0.19662 1.82091 1.0 -2.30251 1.98313 -0.56142 1.0 -0.50340 -1.22275 -0.80844 1.0 1.77113 1.40845 -1.96375 1.0 2.59846 0.62092 0.24509 1.0 0.92301 0.70809 1.75323 1.0 -0.52826 2.32905 0.45154 1.0 1.95452 4.63023 -0.78412 1.0 0.56180 4.04186 -1.31068 1.0 1.91148 3.74975 -2.12028 1.0 3.36786 -2.44250 1.63478 1.0 3.65813 -0.98745 1.03431 1.0 3.90670 -2.28836 0.13592 1.0 1.39812 -3.76189 0.69146 1.0 1.94611 -3.53135 -0.79415 1.0 0.46536 -3.06399 -0.40575 1.0 -1.65462 -2.62956 -1.89823 1.0 -3.12441 -2.10556 -2.25275 1.0 -2.92304 -3.21635 -1.11835 1.0 -2.89157 1.96880 0.22253 1.0 -2.62621 1.33607 -1.22343 1.0 -2.30249 2.88084 -0.95409 1.0 -1.14311 -0.84952 -0.33302 1.0 -1.96606 0.25054 2.87958 1.0 1.73199 2.67666 -0.44912 1.0 -3.35361 -0.92144 0.15549 1.0 1.06231 -1.23843 1.35516 1.0 1.79598 -0.88863 -0.78250 1.0 -0.10860 1.53234 -1.33663 1.0Start by the longer part : adapting your coordinates like in the above format. Because the atoms order should be : C H N O, as defined in the .dat file above, all the C atoms have to be listed first, then all the H atoms, then all the N and finally all the O atoms. You have first to reorder atoms. Then you have to suppress all text and put the occupation number to 1.000 for all atoms. This tedious operation took me exactly 4 minutes and 10 seconds, below is the result :
2.204 1.120 -0.530 1.000 -0.378 0.845 0.905 1.000 -0.531 -0.907 -0.290 1.000 -4.058 0.603 1.110 1.000 -4.163 -0.558 0.332 1.000 -3.020 -1.162 -0.200 1.000 -1.795 -0.576 0.071 1.000 -1.696 0.547 0.832 1.000 -2.809 1.169 1.370 1.000 1.667 -0.067 0.098 1.000 2.089 -1.255 -0.743 1.000 3.611 -1.239 -0.777 1.000 4.032 0.014 -1.318 1.000 -4.900 1.036 1.490 1.000 -5.081 -0.965 0.153 1.000 -3.087 -2.007 -0.766 1.000 -2.720 2.011 1.939 1.000 2.119 -0.174 1.085 1.000 1.691 -1.152 -1.752 1.000 1.736 -2.185 -0.297 1.000 3.970 -2.056 -1.403 1.000 4.006 -1.349 0.233 1.000 3.644 1.878 -1.558 1.000 0.104 1.787 1.510 1.000 -0.216 -1.860 -0.985 1.000 5.081 0.068 -1.939 1.000 1.605 2.185 -0.462 1.000 3.326 1.086 -1.174 1.000 0.285 -0.041 0.232 1.000Reordering the atoms and cleaning the coordinate list is the longer part of the expected 10 minutes. So many molecule formats exist (see Babel, the well named software) that none would satisfy more than 5% of the users... However, ESPOIR could be modified in order to accept directly some formats (.xyz, .m3d, etc).
Now, adapt the rest of the thalid.dat file to your case. The cell parameters, a title, and the U, V, W are in the last .pcr file if you used FULLPROF for structure factor amplitudes extraction. You can do it by a copy-paste fast operation. This will need 2 minutes and 32 seconds.
The thalid.dat file is now :
! title Thalidomide beta ! a, b, c, alpha, beta, gamma 20.679 8.042 14.162 90.0 102.86 90.0 ! space group C 2/C ! lambda, radiation, N of atoms, types of atoms, N of objects, ! "|Fobs|" or patterns, iprint 1.54056 4 29 4 1 1 3 ! U, V, W, step 0.03520 -0.01640 0.01273 3 ! atom names, in 8A4 C H N O ! code for minimal distance contraints 0 ! maximum moves for each type of atom 10.000 10.000 10.000 10.000 ! annealing law, sigma, reject 1.0000 1.0000 0.0050 ! number of events for : print, maximum, save 5000 100000 100000 ! events for restart, rmax, ichi, number of runs 50000 0.400 2 10 ! object type and NPERM for object 1 2 4 ! number of atoms of each type in object 1 13 10 2 4 ! B overall, NOCC, NSPE for object 1 4.0 0 0 ! cell parameters, and x, y, z, occup. for object 1 !Add there the cell parameters ! and the x,y,z, occup. for your object 1 0.0 0.0 0.0 90.000 90.000 90.000 2.204 1.120 -0.530 1.000 -0.378 0.845 0.905 1.000 -0.531 -0.907 -0.290 1.000 -4.058 0.603 1.110 1.000 -4.163 -0.558 0.332 1.000 -3.020 -1.162 -0.200 1.000 -1.795 -0.576 0.071 1.000 -1.696 0.547 0.832 1.000 -2.809 1.169 1.370 1.000 1.667 -0.067 0.098 1.000 2.089 -1.255 -0.743 1.000 3.611 -1.239 -0.777 1.000 4.032 0.014 -1.318 1.000 -4.900 1.036 1.490 1.000 -5.081 -0.965 0.153 1.000 -3.087 -2.007 -0.766 1.000 -2.720 2.011 1.939 1.000 2.119 -0.174 1.085 1.000 1.691 -1.152 -1.752 1.000 1.736 -2.185 -0.297 1.000 3.970 -2.056 -1.403 1.000 4.006 -1.349 0.233 1.000 3.644 1.878 -1.558 1.000 0.104 1.787 1.510 1.000 -0.216 -1.860 -0.985 1.000 5.081 0.068 -1.939 1.000 1.605 2.185 -0.462 1.000 3.326 1.086 -1.174 1.000 0.285 -0.041 0.232 1.000
Finally, for building the .hkl file,
take the .fou file from FULLPROF, rename it as thalid.hkl, remove the first
text line and put a number of hkl = 50 in its place. This will take you
15 seconds maximum.
Up to now, the total time is 6 minutes and 57 seconds (expert time...).
3 - Run ESPOIR
It is recommended to verify soon if you did not make any mistake in your starting model. Prepare a fast run with a small number of Monte Carlo events : 500 for instance. In that way, a first proposal for the molecule location is obtained very quickly (certainly false...) which allows you to see the molecule with RASMOL (click on "view" and then on "structure", select the default .xyz file), and to check it :
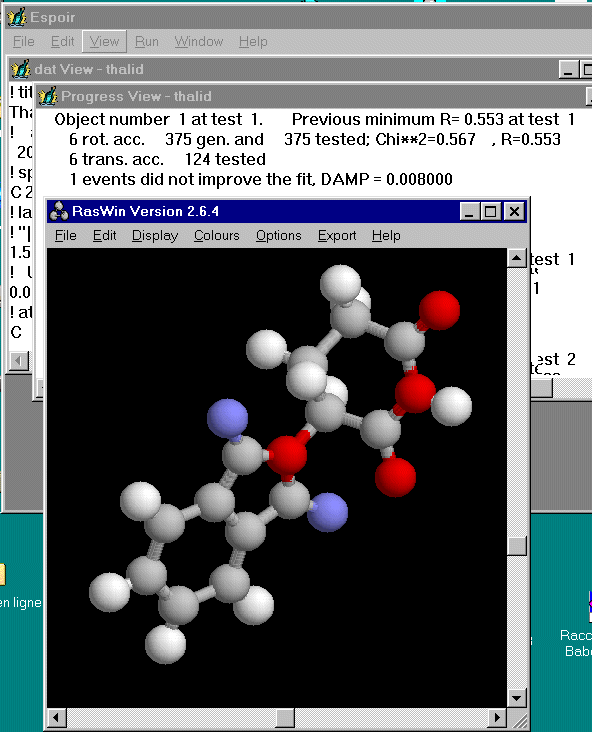
That is looking quite a good starting model ! The molecule checking needed 45 seconds, so that we are now at 7 minutes and 42 seconds.
If you want to let free torsion angles (automatic location of the rotatable bonds) you have to modify the thalid.dat file by a nobt = -3 line, requiring an optional subsequent line in which the maximum rotation angles are given (360° for the whole molecule, and 5° for the torsion angles) :
! object type and NPERM for object 1 -3 4 360. 5.There is obviously one torsion angle in the thalidomide molecule (see the drawing above).
But the best is to start without free torsion angles.
4 - Results
10 tests of 100000 Monte Carlo translations/rotations, without the free torsion angle did not allow to obtain a Rp(F) reliability factor below 38.0%.
10 more tests with the torsion angle allowed to vary led to obtain a Rp(F) value of 16.6%. This is probably close enough to the final structure for attempting now a refinement with restraints.
The whole time for running simultaneously the 2 calculations on a 800 MHz Intel Pentium III PC was 33 minutes.
A total of ~40 minutes for redetermining the structure of b-thalidomide ;-).
If you have already CHIME installed, you may see the ESPOIR proposition in 3D below :
If not, see the .gif file :
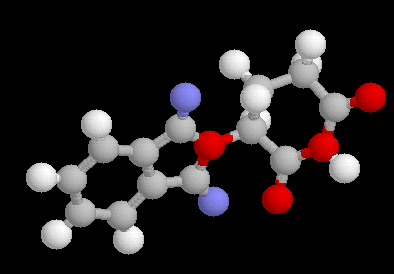
If both results had been negative, then other starting parameters may have to be selected (more Monte Carlo events, more independent tests, different pseudo-annealing law, different maximum value for the torsion angle rotation, etc) implying to add time to the files setup.
Both data sets and results are viewable by downloading thalid.zip.
Best wishes !
October 2000
Copyright © 2000 - Armel Le Bail